Abstract
Background: The therapeutic effect of DNA-hypomethylating agents (HMAs) in AML/MDS is discussed to be via its effects on aberrant gene silencing by reactivation (e.g. through promoter hypomethylation). While this has been broadly studied in cell line models, only very few studies have addressed the global effects of HMAs in primary blasts serially isolated from AML patients (pts) undergoing HMA treatment (Claus et al., Leuk. Res. 2013, Klco et al., Blood 2013, Welch et al., N. Engl. J. Med. 2016). We therefore conducted prospective serial methylome and transcriptome analyses on AML blasts from pts of the DECIDER trial (NCT00867672), hypothesizing that both random and non-random effects of the HMA may be observed in vivo.
Patients, Materials and Methods: Of a total of 200 newly diagnosed AML pts included into the DECIDER randomized phase II trial (Decitabine/DAC treatment, 20 mg/m2 intravenous 1-hour infusion over 5 days, with add-on drugs Valproic acid and/or ATRA added at day 6; Grishina et al., BMC Cancer 2015), serially obtained peripheral blood (pb) samples from a total of 28 pts yielded sufficient numbers of purified blasts at 3 timepoints (days 0, 8 and 15 from DAC treatment start) to allow a "triplet analysis" of these matched samples. Baseline pt characteristics: median WBC 11,900/µl (range 1,200 - 53,800), median pb blasts 37.5% (range 1% - 93%). Blasts were sorted using anti-CD34, CD117 MACS microbeads, respectively (median purity >90%). Methylomes were obtained using Infinium Human Methylation 450 BeadChip arrays (Illumina). For expression analyses, GeneChip Human Gene 2.0 ST arrays were used. A linear-model based approach was used to identify the differentially methylated CpGs and expressed genes post vs. prior to treatment.
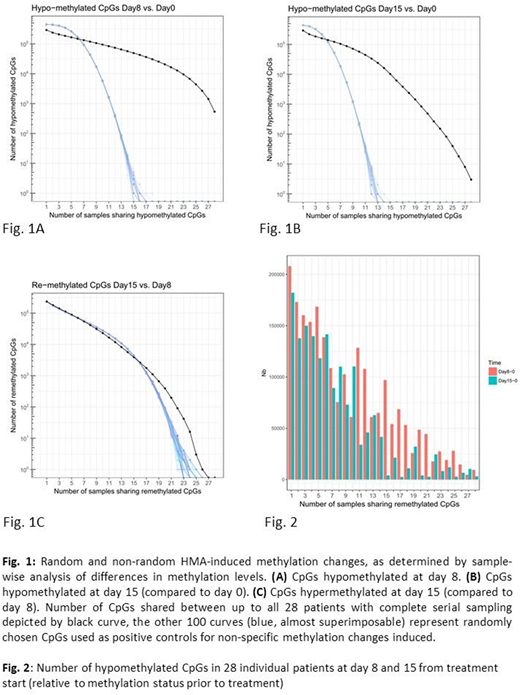
Results: To address in vivo methylation changes occurring at day 8 and 15 from DAC treatment start, complete "triplets" of DNA preparations (thus from purified pb blasts of all 3 time points) were interrogated. Significant hypomethylation at day 8 (Δβ<-0.1, FDR<0.05) was achieved for 69,384 CpGs (15% of all 456,789 CpGs). Of these, 536 CpGs (representing 227 genes) became hypomethylated in all 28 pts (Fig. 1A, black curve). By Gene Ontology (GO) terms, there was strong enrichment for genes involved in adhesion; this included a number of genes with proven or putative tumor suppressor function, several of them known to be hypermethylated in AML or other malignancies, such as CDH13, FAT1 and CYP26B1. By day 15, 40129 CpGs (58% of the 69,384 CpGs) became at least partially remethylated (Fig. 2). The median methylation of all CpGs was thus reduced from 0.91 pre-treatment to 0.76 at day 8, and increased to 0.81 at day 15. We next asked whether the CpG methylation changes at different timepoints were random or specific. Compared to 100 randomly chosen sets of CpGs (positive controls for non-specific methylation changes), the number of CpGs commonly hypomethylated (Δβ <-0.1) at day 8 (vs. day 0) across up to 28 pts was much higher, as shown in Fig. 1A (p=3.78e-42), indicating non-random hypomethylation. Similar results were obtained for CpGs hypomethylated at day 15 (vs. day 0, Fig. 1B, p=2.61e-25). In contrast, CpGs remethylated at day 15 (vs. day 8) were similar in numbers to the random controls (Fig. 1C, p=0.42), thus indicating random remethylation. We next investigated whether hypomethylation at day 8 was associated with changes in gene expression. For 12 of the 28 pts, blast RNA yield was sufficient to allow for combined methylation and expression studies both prior to treatment and at day 8. Regarding expression induction upon HMA treatment, a total of 870 genes showed a significant inverse correlation between expression and DNA methylation, thus being both induced and hypomethylated (with, however, limited overall fold changes). GO analysis revealed enrichment for immune response genes.
Conclusions: In our study, a subset of genes was specifically targeted by hypomethylation in all pts, arguing against a completely random effect of this treatment on the methylome. Only a limited number of hypomethylating events was associated with gene reactivation. A better understanding of hypo- and remethylation kinetics in vivo may aid in schedule optimization of HMA therapy. Correlative studies on hypomethylation kinetics and treatment outcome in the DECIDER trial are therefore ongoing, as well as serial methylome analyses in the EORTC trial AML21 ("inDACtion vs. induction", NCT02172872).
Salih:Several patent applications: Patents & Royalties: e.g. EP3064507A1. Ganser:Novartis: Membership on an entity's Board of Directors or advisory committees. Döhner:Jazz: Consultancy, Honoraria; Celator: Consultancy, Honoraria; Sunesis: Consultancy, Honoraria, Research Funding; Jazz: Consultancy, Honoraria; Celator: Consultancy, Honoraria; Bristol Myers Squibb: Research Funding; Bristol Myers Squibb: Research Funding; Amgen: Consultancy, Honoraria; Celgene: Consultancy, Honoraria, Research Funding; Sunesis: Consultancy, Honoraria, Research Funding; Celgene: Consultancy, Honoraria, Research Funding; AROG Pharmaceuticals: Research Funding; Astellas: Consultancy, Honoraria; Astex Pharmaceuticals: Consultancy, Honoraria; AROG Pharmaceuticals: Research Funding; Astellas: Consultancy, Honoraria; Amgen: Consultancy, Honoraria; Novartis: Consultancy, Honoraria, Research Funding; AbbVie: Consultancy, Honoraria; Agios: Consultancy, Honoraria; Seattle Genetics: Consultancy, Honoraria; Janssen: Consultancy, Honoraria; Janssen: Consultancy, Honoraria; Pfizer: Research Funding; Novartis: Consultancy, Honoraria, Research Funding; Pfizer: Research Funding; Seattle Genetics: Consultancy, Honoraria; AbbVie: Consultancy, Honoraria; Agios: Consultancy, Honoraria; Astex Pharmaceuticals: Consultancy, Honoraria. Lübbert:Cheplapharm: Other: Study drug; Janssen: Honoraria, Research Funding; TEVA: Other: Study drug; Celgene: Other: Travel Support.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal